| Issue | Solutions |
|---|---|
| Over-fitting | - Model ensembles - 3-way cross validation - Pruning inputs |
| Black boxes models | - Plot partial derivatives - Feature importance |
| Computationally expensive | - Tensorflow - GPU processing |
Tools and Guidance for Applying Neural Networks to Eddy Covariance Data
Eddy Covariance
Semi-continuous, ecosystem-scale energy, water, and trace gas fluxes.
- Noisy, voluminous data sets
- Frequent gaps
- Observational bias
- Well suited for machine learning!

Burns Bog EC Station
Delta, BC
Neural Networks
Universal approximators: can map any continuous function to an arbitrary degree accuracy.
- With enough hidden nodes, will fit any pattern in a dataset
- Care must be taken to ensure the patterns are real
- Early stopping can help prevent over-fitting
- Well suited for non-linear, multi-variate response functions
- Capable of interpolation and extrapolation
Commonly Cited Limitations
Objective
Provide a framework for applying NN models to EC data for descriptive analysis and inferential modelling.
- The github repository linked here has functional examples that can be used to setup NN models.
- Runs in Python and Tensorflow
- GPU support not required
- But will decrease processing times
- GPU support not required
- Runs in Python and Tensorflow
Example Data
Burns Bog EC station
- Harvested peatland undergoing active restoration
- 8+ years of flux data

Training Procedures
- Larger ensemble = more robust model
- N \(\leq\) 10 for data exploration/pruning
- Three way cross-validation
- Train/validate/test
- Early Stopping: after e epochs
- e = 2 for pruning stage
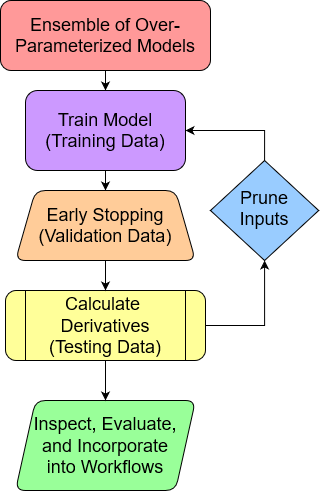
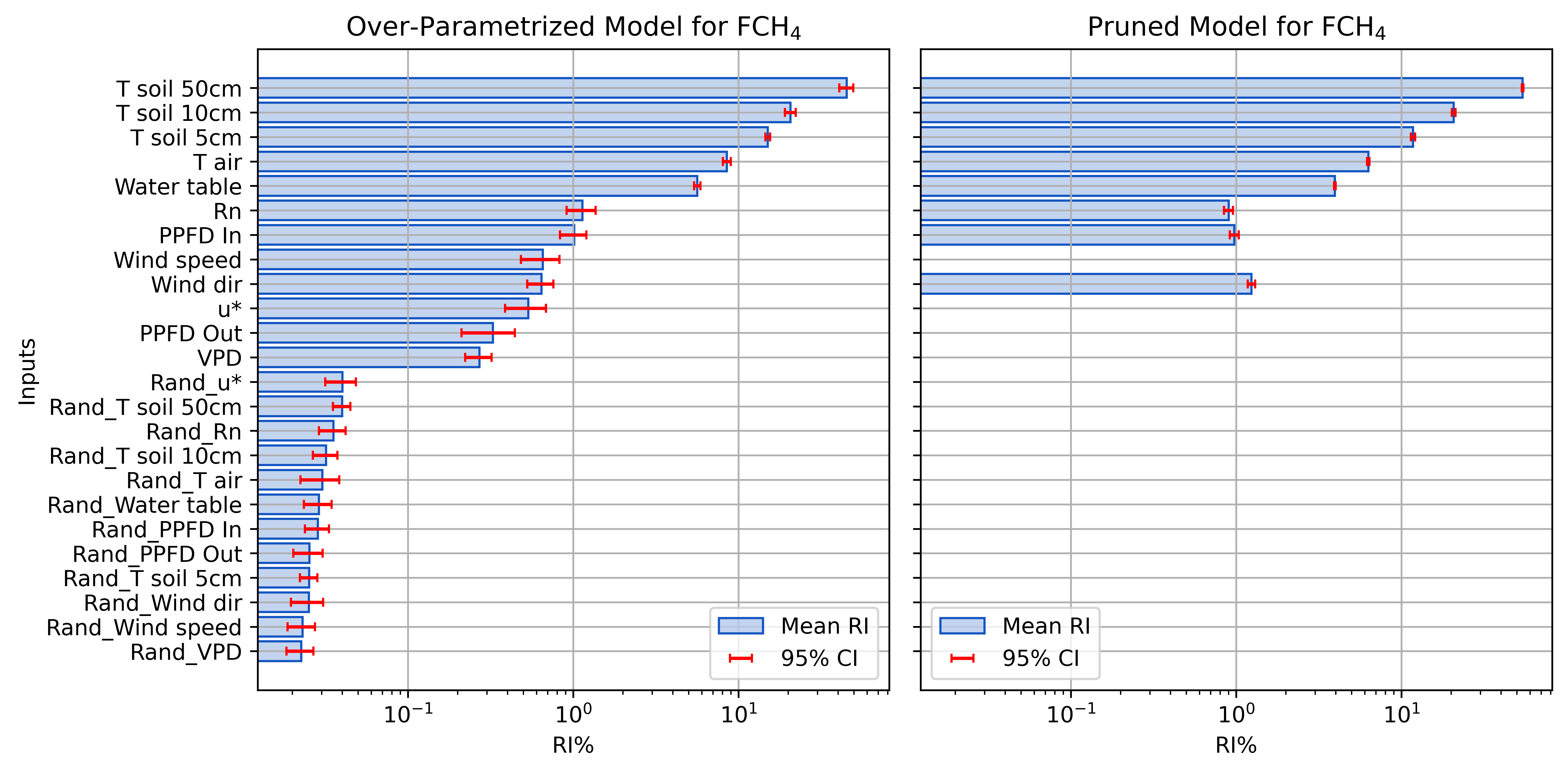
Pruning Inputs
Calculate partial first derivative of the output with respect to each input over test data domain.
- Relative Influence (RI) of inputs
- Normalized sum of squared derivatives (SSD)
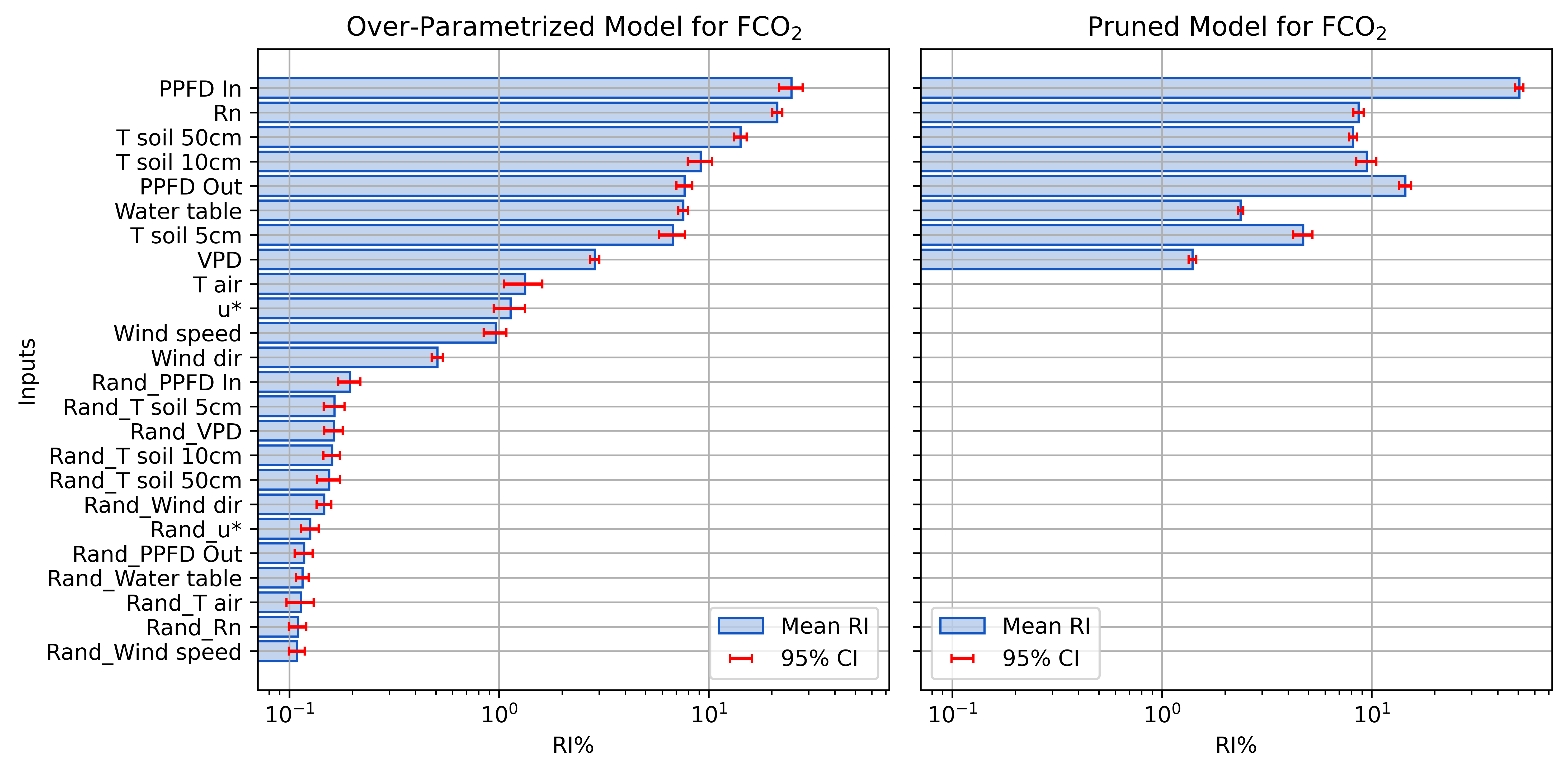
- Iteratively remove inputs with RI below a threshold
- Use random noise to determine threshold
- e.g., randomly shuffled copy of each input
- Use random noise to determine threshold
Before and After Pruning FCO2
The Final Model
Once pruning is complete, re-train the final production level model, excluding the random scalars
- Increase the ensemble size (e.g., N \(\geq\) 30)
- Increase early stopping (e) criteria (e.g., e = 10)
- Larger e drastically increases training time
- Increase early stopping (e) criteria (e.g., e = 10)
- Plot the model derivatives as a final check
- If derivatives look implausible
- Adjust inputs/parameters and try again
- If derivatives look implausible
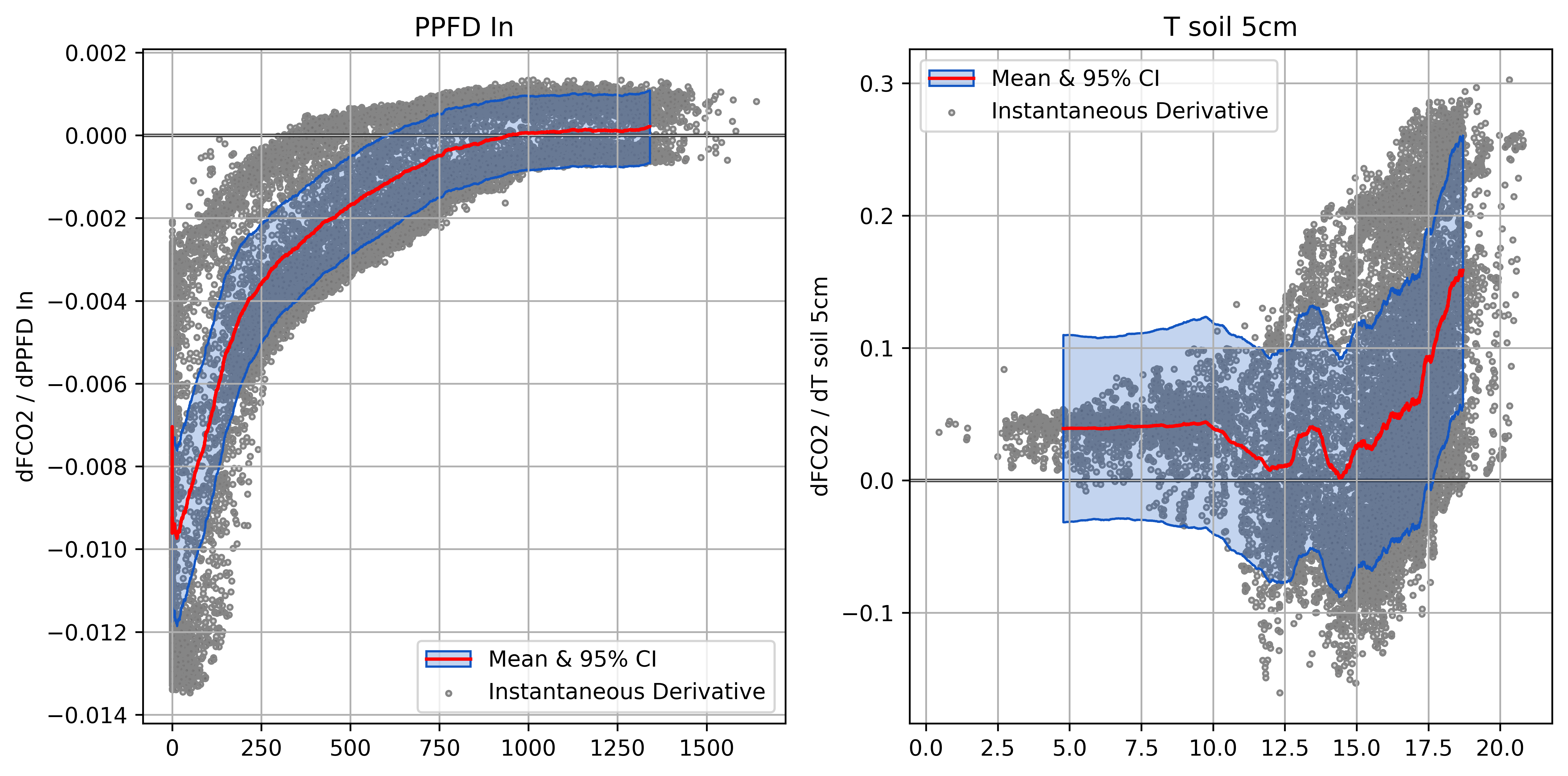
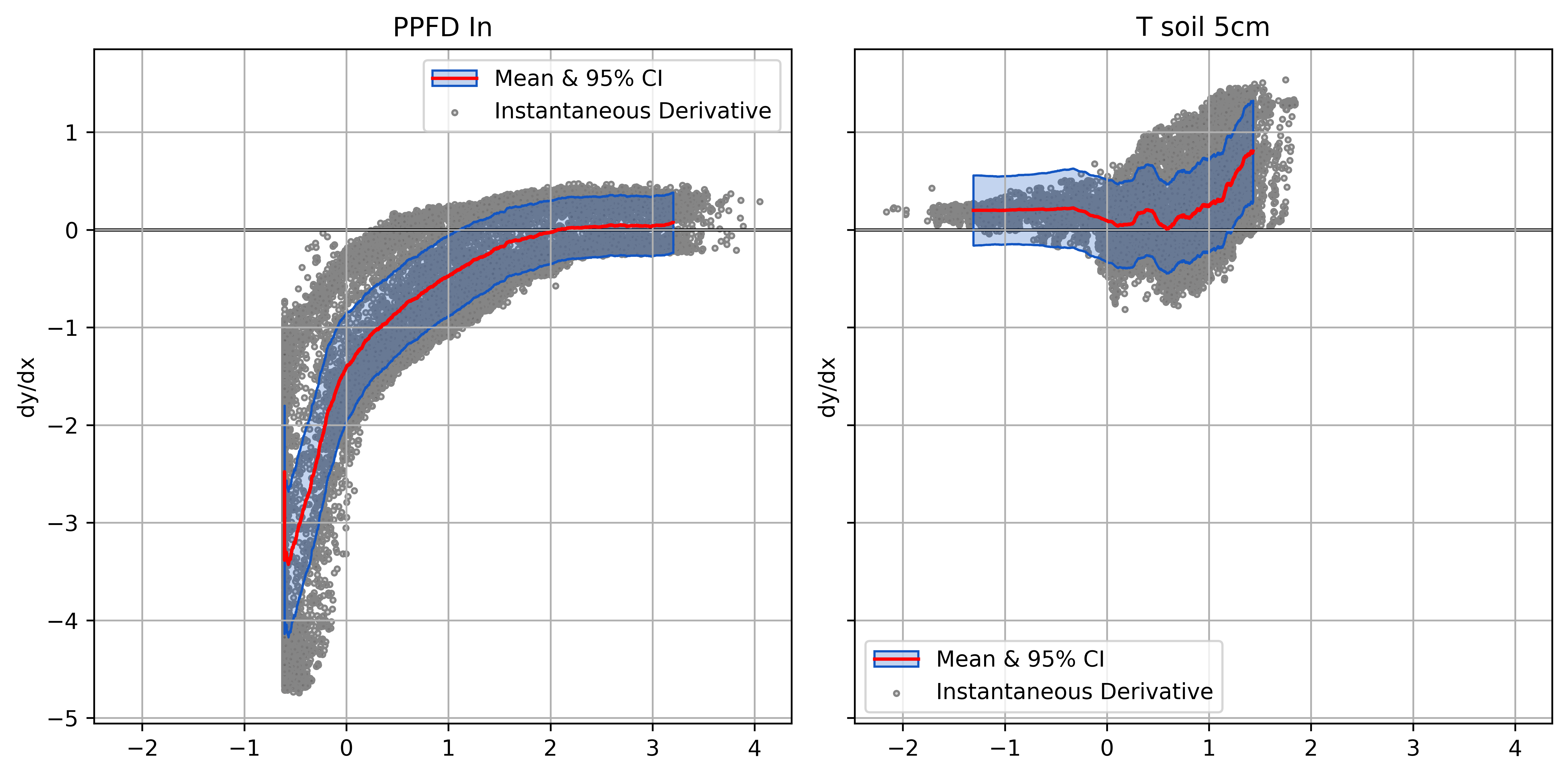
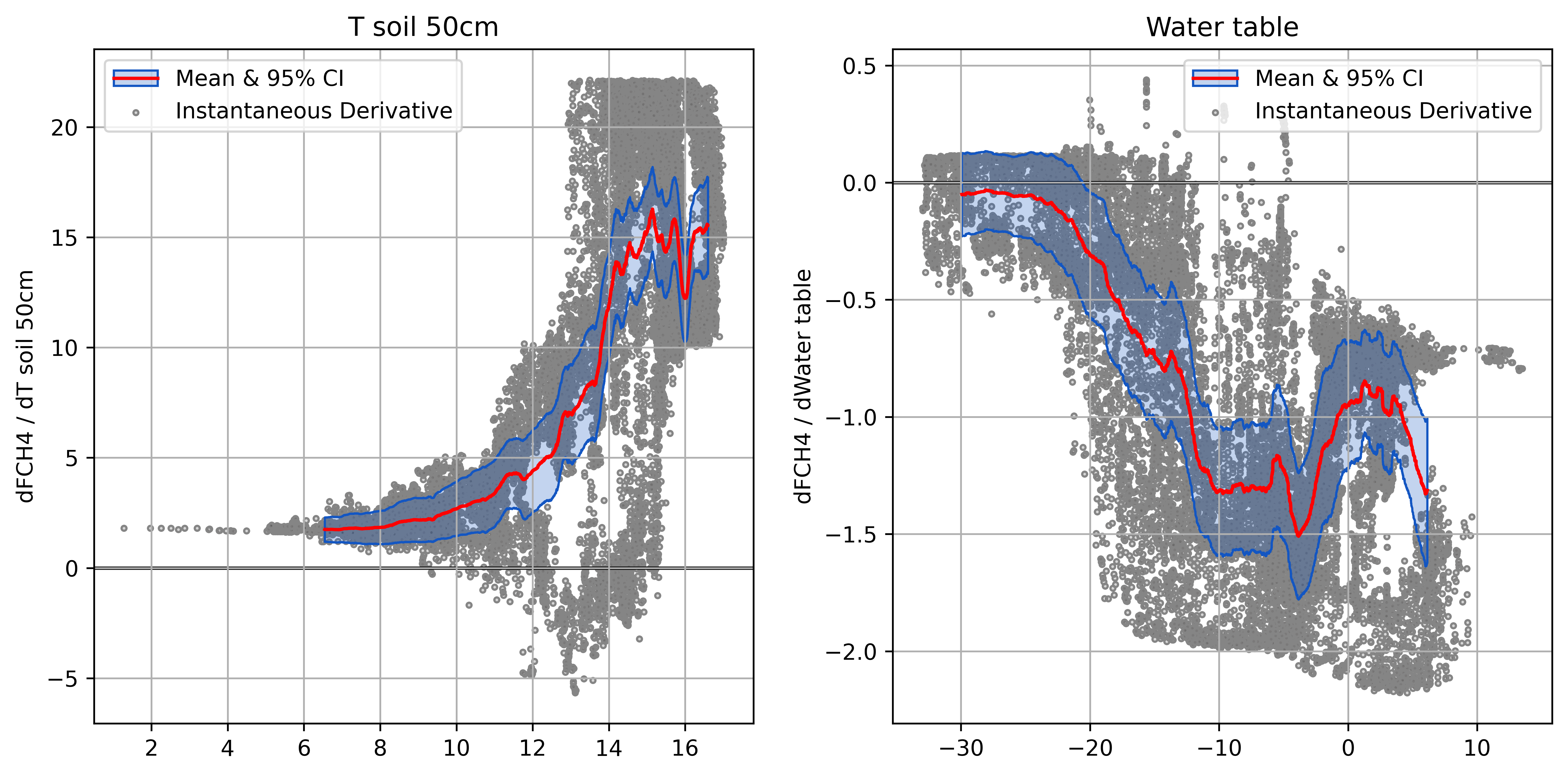
Plotting Derivatives
Helps ensure model responses are physically plausible
- An essential step and key advantage of NN models
- Raw derivatives show response in natural units
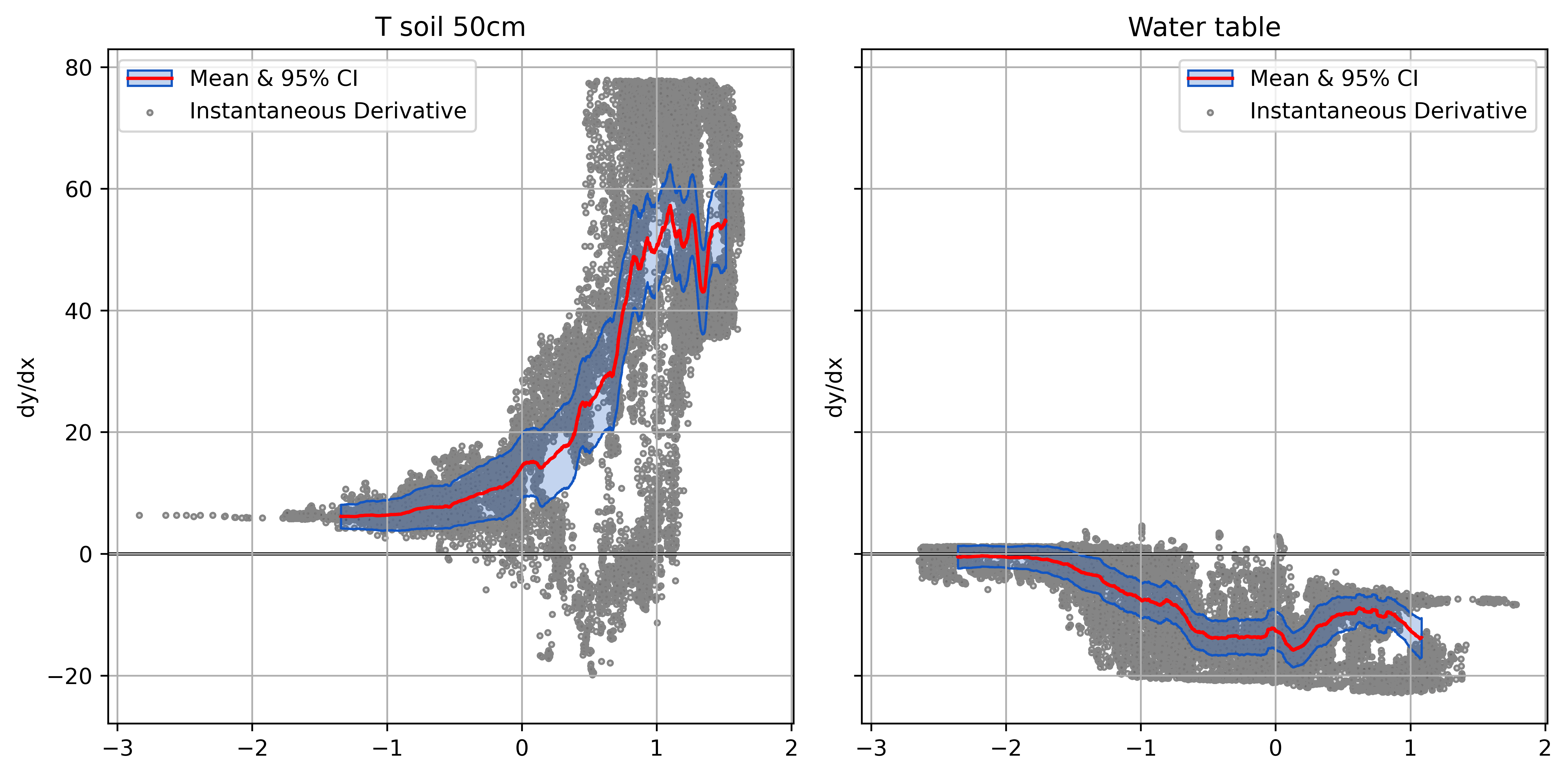
- Normalized derivatives scaled by input variance
- Relative input effects on common scale
- What the model “sees”
Partial Derivatives of FCO2
Normalized Derivatives of FCO2
Model Performance FCO2
Plot the model outputs and validation metrics calculated with the test data.
| Metric | Score |
|---|---|
| RMSE | 0.64 \(\mu mol\) \(m^{-2}s^{-1}\) |
| r2 | 0.88 |
Before and After Pruning FCH4
Partial Derivatives of FCH4
Normalized Derivatives of FCH4
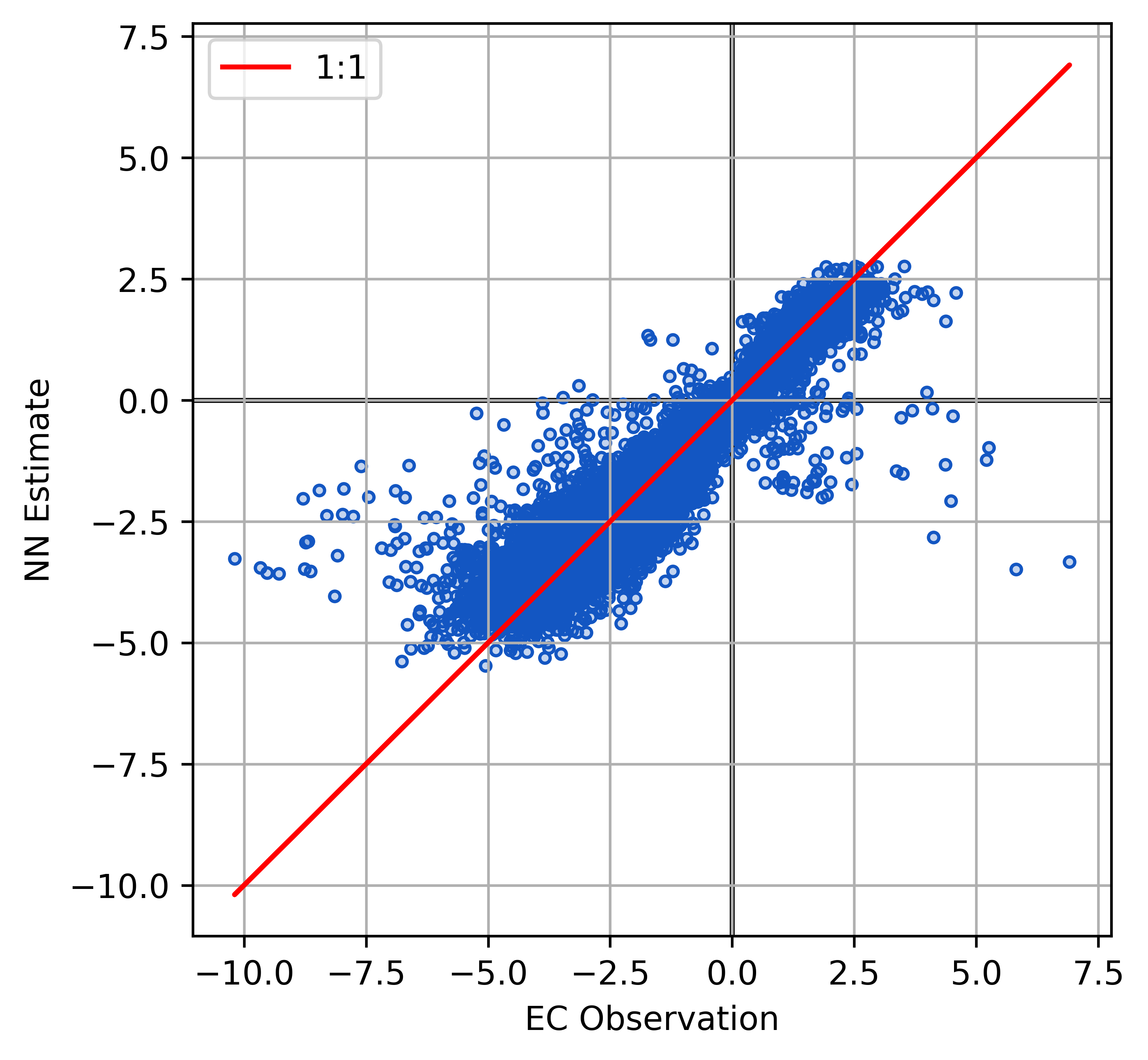
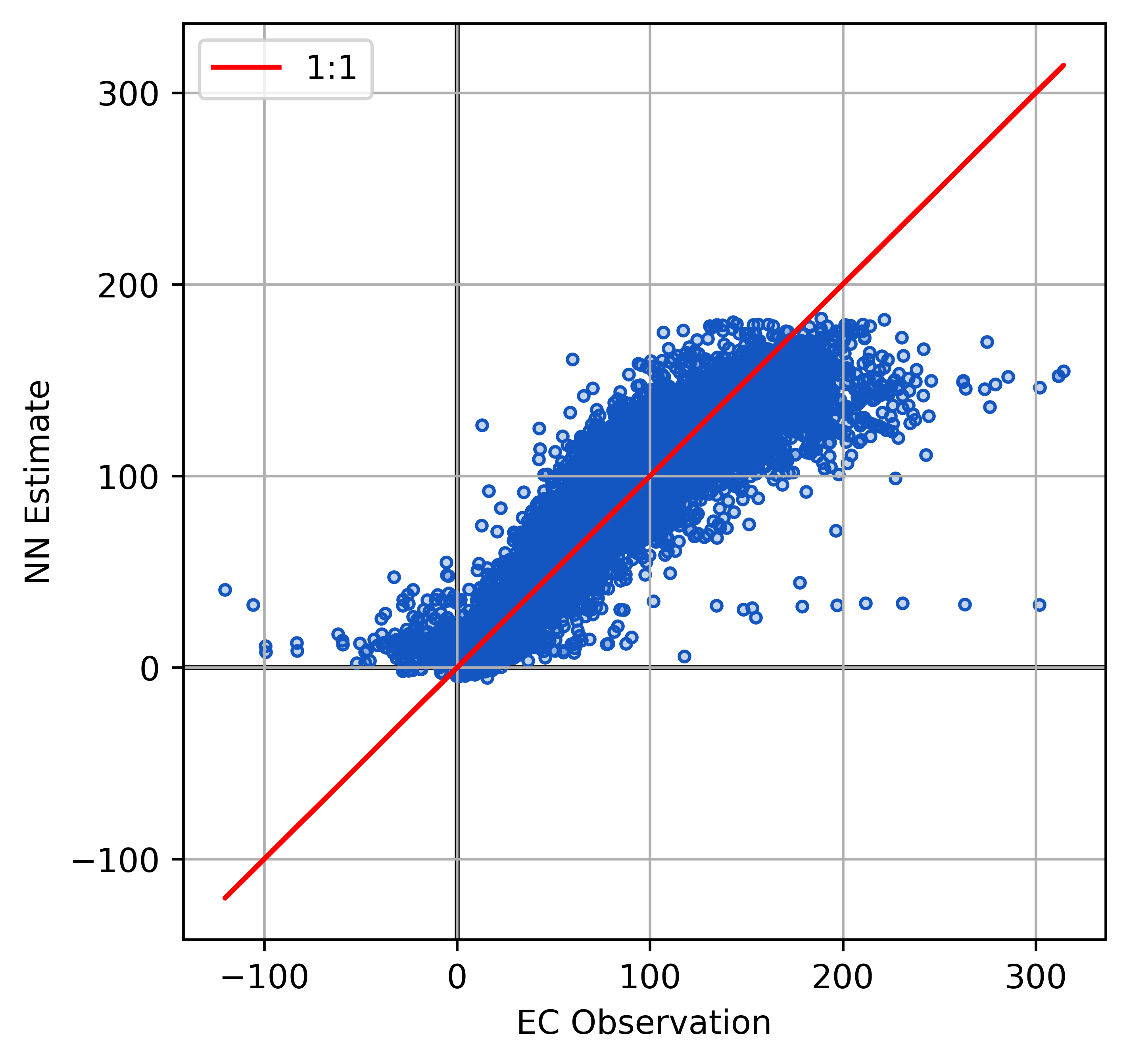
Model Performance FCH4
Plot the model outputs and validation metrics calculated with the test data.
| Metric | Score |
|---|---|
| RMSE | 17.17 \(nmol\) \(m^{-2}s^{-1}\) |
| r2 | 0.89 |
Next Steps
- Custom NN architecture: Separating input layers may allow us partition fluxes.
- e.g., FCO2 into GPP and ER
- Flux footprints: map response to spatial heterogenity
- Upscaling: in space and time
- u* filtering: partial derivatives could identify u* thresholds
- Compare to process based models (e.g., CLASSIC)
Thank You
Questions?